Research Overview
Regulation of Gene Expression by Cyclin-dependent Kinases 11 and 12 (CDK11 and CDK12)
RNA Polymerase II (RNAPII) directs the transcription of protein-coding genes in a process consisting of initiation, elongation and termination. It contains an unstructured C-terminal domain (CTD) with 52 repeats of evolutionarily conserved heptapeptide YSPTSPS, where individual serines (Ser) (Ser2, Ser5, Ser7), threonine 4 (Thr4) and tyrosine 1 (Tyr1) are phosphorylated. The CTD is necessary for the regulation of the transcription cycle and for coupling transcription with co-transcriptional processing. P-Ser2 is a marker of active elongation and plays a stimulatory role in co-transcriptional mRNA processing via the recruitment of splicing, cleavage and polyadenylation factors.
Several kinases, including well-studied CDK7 and CDK9, phosphorylate the CTD of RNAPII and other transcription factors to regulate individual steps of transcription and co-transcriptional processing. Our work has indicated important roles of other less-studied CDKs, including CDK11 and CDK12, in the regulation of transcription elongation and co-transcriptional mRNA processing. Notably, all the known P-Ser2 CTD and elongation-associated kinases are dysregulated in human diseases, including cancer, and their inhibition is considered to be a very promising strategy for fighting various diseases.
Ongoing work in my lab will further explore the roles of CDK11 and CDK12 in the regulation of RNAPII-directed gene expression, particularly in transcription elongation and mRNA-processing. We use genomic, proteomic and biochemical approaches to achieve these goals.
Regulation of transcriptional elongation and genome stability by CDK12
CDK12 phosphorylates the CTD of RNAPII and other substrates to regulate the elongation of transcription. We found that CDK12 maintains genome stability via regulating the transcription elongation of key DNA damage response genes including BRCA1, ATR, ATM, FANCD2 and FANCI (Blazek et al, G&D, 2011). CDK12 kinase activity is also needed for the optimal transcription of core DNA replication genes and thus for G1/S cell cycle progression (Chirackal Manavalan et al., EMBO reports, 2019). We propose that by regulating gene-specific expression, CDK12 has pleiotropic effects on genome stability in cell lines and various tumors (Pilarova et al, NAR Cancer, 2020). Consistent with this, comprehensive genomic analyses of patient samples revealed CDK12 to be disregulated in various types of cancer. We showed that CDK12 mutations in high-grade serous ovarian carcinoma prevent the formation or activation of the CDK12/CYCK complex, rendering the kinase inactive (Ekumi et al, NAR, 2015).
Ongoing research is focusing on the molecular mechanism of CDK12-dependent gene expression including the identification of novel substrates. We are also interested in exploiting CDK12 vulnerabilities in cancer treatment.
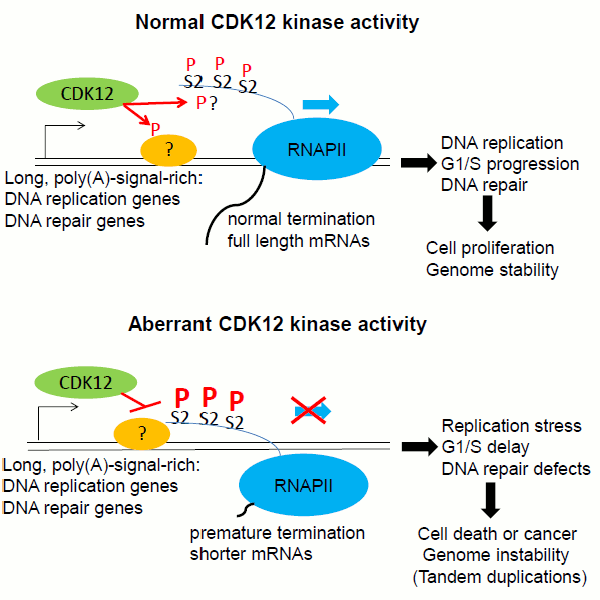
(Chirackal Manavalan et al, EMBO reports, 2019)
Roles of CDK11 in gene-specific expression
CDK11 is a little-studied, but essential kinase, and is believed to play a role in RNA polymerase II (RNAPII)-directed transcription and co-transcriptional mRNA-processing. However, so far its specific roles in gene expression have been unknown. Importantly, numerous recent studies have demonstrated many cancers to be dependent on CDK11 activity.
We have found that CDK11 is required for the transcription of replication-dependent histone genes. The kinase binds FLASH protein, and with the contribution of replication-dependent histone mRNAs, recruits CDK11 to the middle of the histone genes, where it is needed for Ser2 phosphorylation and their optimal elongation and 3´ end processing (Gajduskova et al, Nature Structural & Molecular Biology, 2020).
Ongoing research in my lab is focusing on uncovering the molecular mechanisms and cellular functions of this enigmatic kinase. We are also interested in exploiting CDK11 vulnerabilities in human diseases.
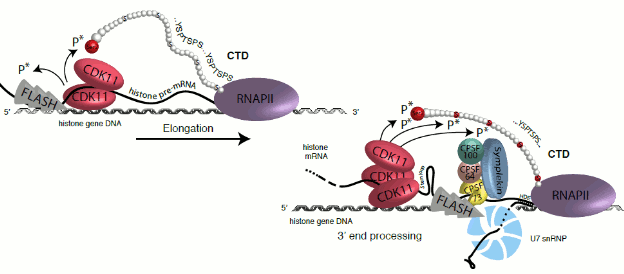
(Gajduskova et al, Nature Structural & Molecular Biology, 2020)
Summary of Main Areas of Research and Approaches
- Roles of CDK11 and CDK12 in the regulation of gene expression
- Roles of CDK12 in the maintenance of genome stability
- Roles of CDK12 in the regulation of cell cycle progression
- Phosphorylation of the C-terminal domain of RNA polymerase II and other substrates by CDK11 and CDK12
- The deregulation of CDK11 and CDK12 in the onset and development of cancer
- Proteomics, Genomics, Chemical Biology